Rhomboid Bifiltration (multicover)
The Rhomboid Bifiltration is a topologically equivalent bifiltration to the multicover bifiltration, introduced in [Computing the Multicover Bifiltration], whose code is available here.
import numpy as np
import gudhi as gd
import multipers as mp
import matplotlib.pyplot as plt
[KeOps] Warning : CUDA libraries not found or could not be loaded; Switching to CPU only.
Definition
Let \(P\) be a point cloud in some metric space \((X,d)\). The multicover bifiltration is the bifiltration \(F\) indexed over \(\mathbb{R} \times \mathbb{N}^{\mathrm{op}}\), defined as follows:
Note that this filtration is not 1-critical/free. The Rhomboid Tiling bifiltration is a topologically equivalent 1-critical bifiltration, that we do not develop here.
We identify \(\mathbb N^{\mathrm{op}}\) with \(-\mathbb N\).
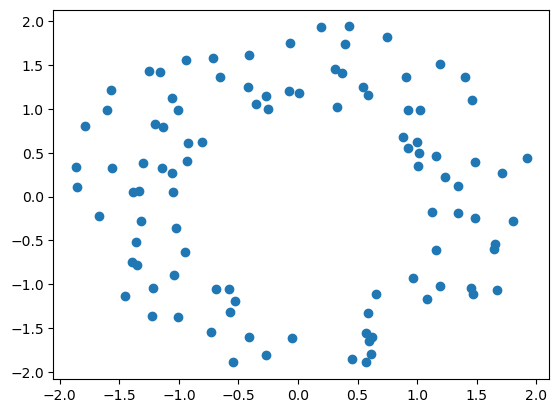
A first example
np.random.seed(0)
x = mp.data.noisy_annulus(100,0, dim=2) # dataset
plt.scatter(*x.T)
k_max = 30 # the maximum of ball intersection to look at
degree=1 # homological degree
s = mp.filtrations.RhomboidBifiltration(x, k_max=k_max, degree=degree)
len(s)
715517
This filtration is very large in general ; before computing an invariant it is generally recommended to simplify it, via a sequence of squeezes or minimal presentations.
multicover = (s
.grid_squeeze(
# resolution=200, # coarsens the filtration grid
# strategy="regular_closest",
threshold_min=[0, -k_max], # thresholds the filtration values
threshold_max=[4, 0],
)
.minpres(degree) # mpfree
.astype(vineyard=True) # for mma
)
len(multicover)
59380
Hilbert Function
The signed barcodes of this filtration are not very readable
sm, = mp.signed_measure(multicover, degree=degree, plot=True);
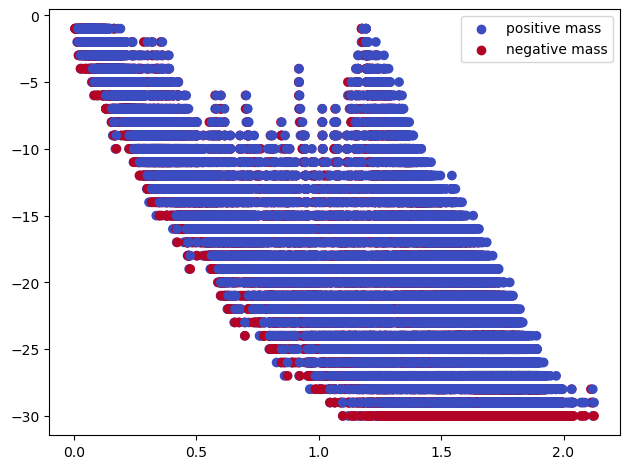
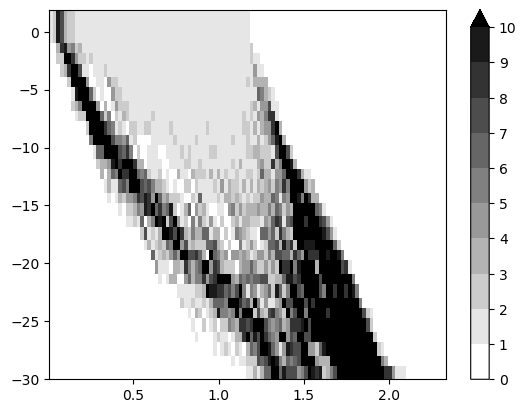
The induced Hilbert function suggest a significant cycle
mp.point_measure.integrate_measure(*sm, plot=True);
Module approximation
mod = mp.module_approximation(multicover)
mod.plot()
/var/folders/w6/5k5w13s94bq0dfsx2xzqxcsh0000gn/T/ipykernel_7492/2670604959.py:1: UserWarning: (copy warning) Got a squeezed input.
mod = mp.module_approximation(multicover)
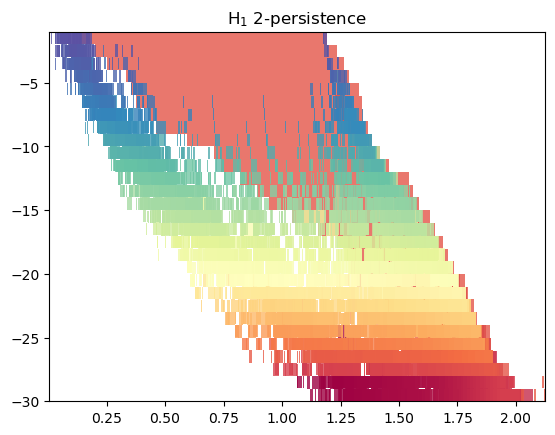
This is a bit bloated, so we can just plot the summand larger than a given threshold. We clearly recover the inital cycle here.
mod.plot(min_persistence = .1)
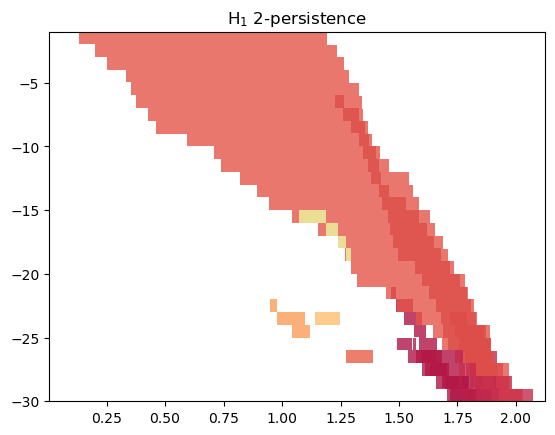
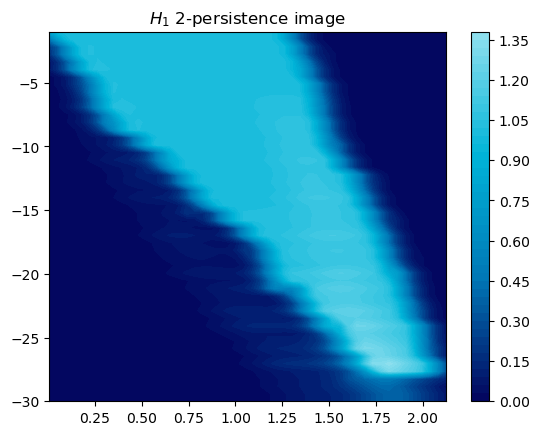
Another alternative is to look at a representation of MMA, which weights the summands w.r.t. their “size”.
mod.representation(plot=True, degrees=[1]);
Scaling
As far as I tested, this filtration doesn’t scale much more than with a few hundreds of points.