Delaunay Lowerstar filtrations
This filtration was introduced by [Delaunay Bifiltrations of Functions on Point Clouds, Alonso et al], whose code is available here.
This bifiltration is very similar to the RipsLowerstar multifiltrations, and is more adapted to low-dimensional Euclidean data.
Definition
Let \(X\) be a point cloud in \(\mathbb R^n\), and \(f\colon X\to \mathbb R\).
The CěchLowerstar bifiltration \(F\) of the point cloud \(X\) , with lowerstar function \(f\) is the bifiltration over \(\mathbb R_+\times \mathbb R\), given by:
The DelaunayLowerstar bifiltration is a 1-critical bifiltration that is topologically equivalent to this one.
An example
import multipers as mp
import matplotlib.pyplot as plt
[KeOps] Warning : CUDA libraries not found or could not be loaded; Switching to CPU only.
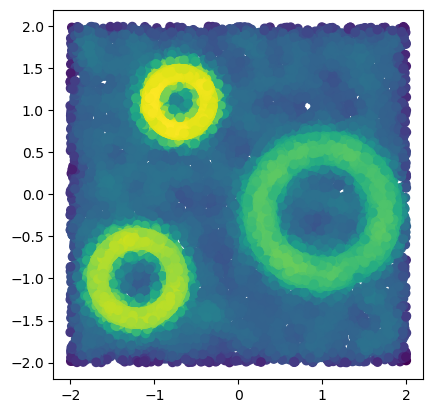
x = mp.data.three_annulus(10_000, 10_000)
f = - mp.filtrations.density.KDE(bandwidth=.1, return_log=True).fit(x).score_samples(x)
plt.scatter(*x.T, c = -f)
plt.gca().set_aspect(1)
The bifiltration can then be obtained by:
s = mp.filtrations.DelaunayLowerstar(points=x, function=f)
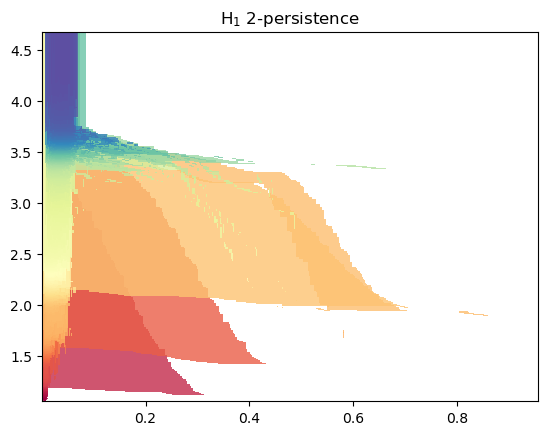
If only a given homological degree is necessary, this construction can made faster:
s = mp.filtrations.DelaunayLowerstar(points=x, function=f, reduce_degree=1)
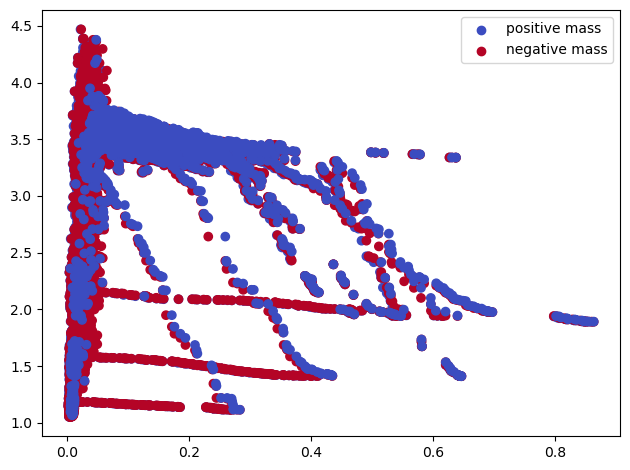
mp.module_approximation(s).plot()
/var/folders/w6/5k5w13s94bq0dfsx2xzqxcsh0000gn/T/ipykernel_8190/1891258844.py:1: UserWarning: (copy warning) Got a non-vine slicer as an input. Use `vineyard=True` to remove this copy.
mp.module_approximation(s).plot()
Autodiff
This bifiltration is not autodiff per se, but can be weakenned into a flag complex to be autodiff w.r.t. the initial point cloud, and the function \(f\).
The gradient is only guaranteed almost everywhere, by [Differentiability and Optimization of Multiparameter Persistent Homology].
import torch
y = torch.from_numpy(x).requires_grad_(True)
f = - mp.filtrations.density.KDE(bandwidth=.1, return_log=True).fit(y).score_samples(y)
st = mp.filtrations.DelaunayLowerstar(points=y, function=f, flagify=True)
Non-problematic operations should preserve gradient
s = mp.Slicer(st).minpres(1)
and invariants should preserve the gradient as well.
(pts,w), = mp.signed_measure(
s,
degree=1,
invariant="hilbert",
plot=True
)
pts.requires_grad # Should be True
True